
When
2022 –– 2024
Sector
Startup, Biotech
Role
Frontend Developer
For
ProteraStage
MVP -> Production
Plaform
Web
Tech stack
Vite, React, TypeScript, Redux-Toolkit, RxJS, Tailwind, Motion (Prev framer-motion), Figma, Jira, Notion
I joined Protera in early 2022 to help build something new: madi, a web platform for protein engineering. The company already had powerful in-house AI models capable of optimizing proteins, but nothing had been productized yet. The mission was to turn all that internal tech into a SaaS product that real users scientists, researchers, engineers could actually use.
madi™ was designed from the ground up to help users design, analyze, and optimize proteins end-to-end. It had to be smart, scalable, and simple enough to not scare away scientists who weren't used to software products but powerful enough to respect their domain.
Not your average SaaS
While on paper it looked like a typical SaaS, the reality was way more complex. The feature scope was still evolving, and the UI had to support heavy data visualizations, custom dashboards, and interactive molecular renderings.
I chose to go with a SPA architecture built with Vite, React, TypeScript, Redux Toolkit, and RxJS. That gave us the flexibility to shape the platform however we needed from routing to state management to handling niche frontend behaviors. Being free from rigid frameworks was key for fast iteration.
This was biotech and in biotech, you often don't have a library ready for what you need. You invent. You adapt. Our users were protein engineers, lab scientists, ML researchers people with zero patience for flaky UIs or meaningless simplifications. We had to translate deep scientific concepts into clean, interactive tools. It wasn't easy, but it was exciting.
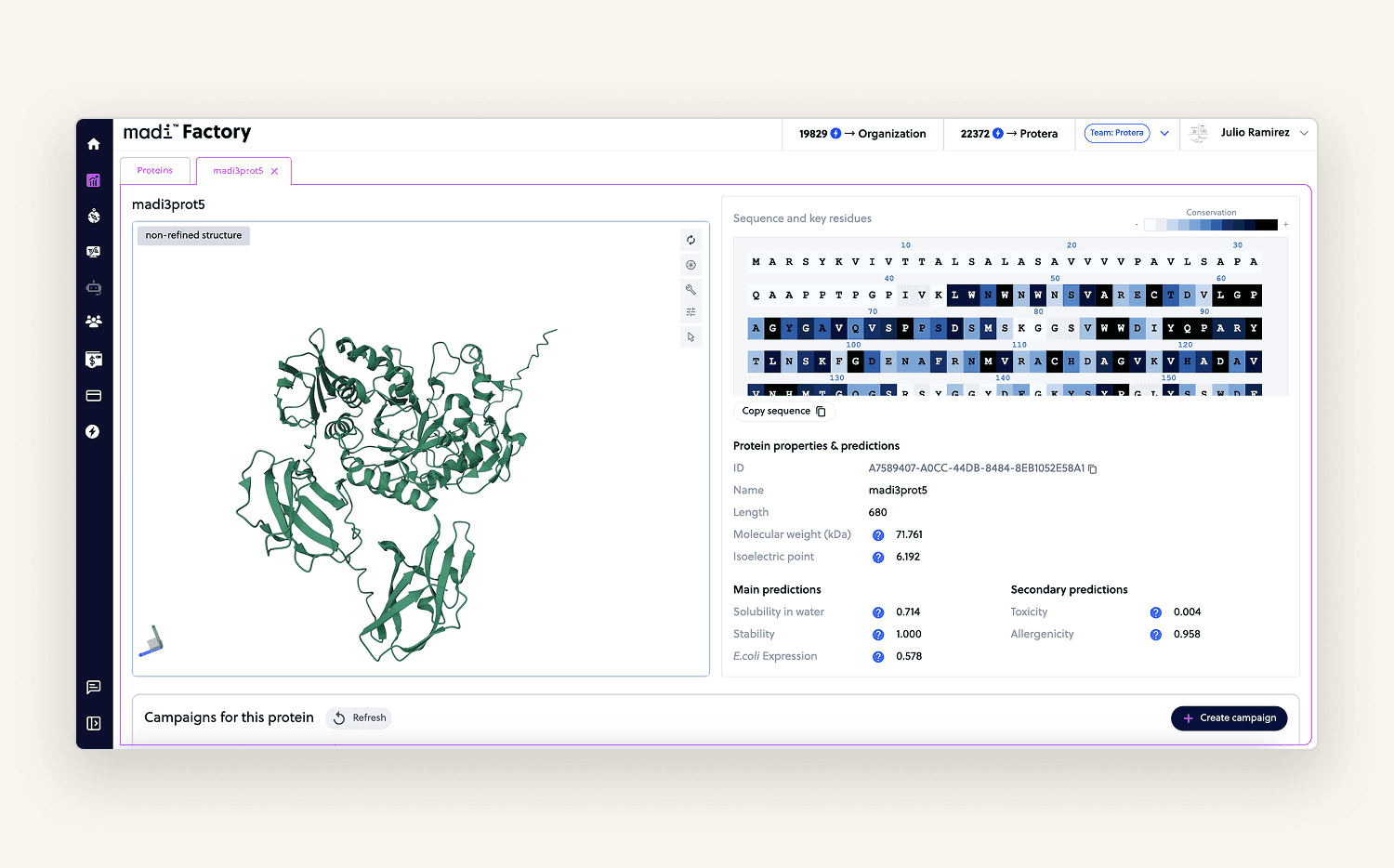
Protein Factory
This was the core feature of the platform. Users could upload a protein (as an amino acid sequence), visualize it, and optimize it using our internal AI models.
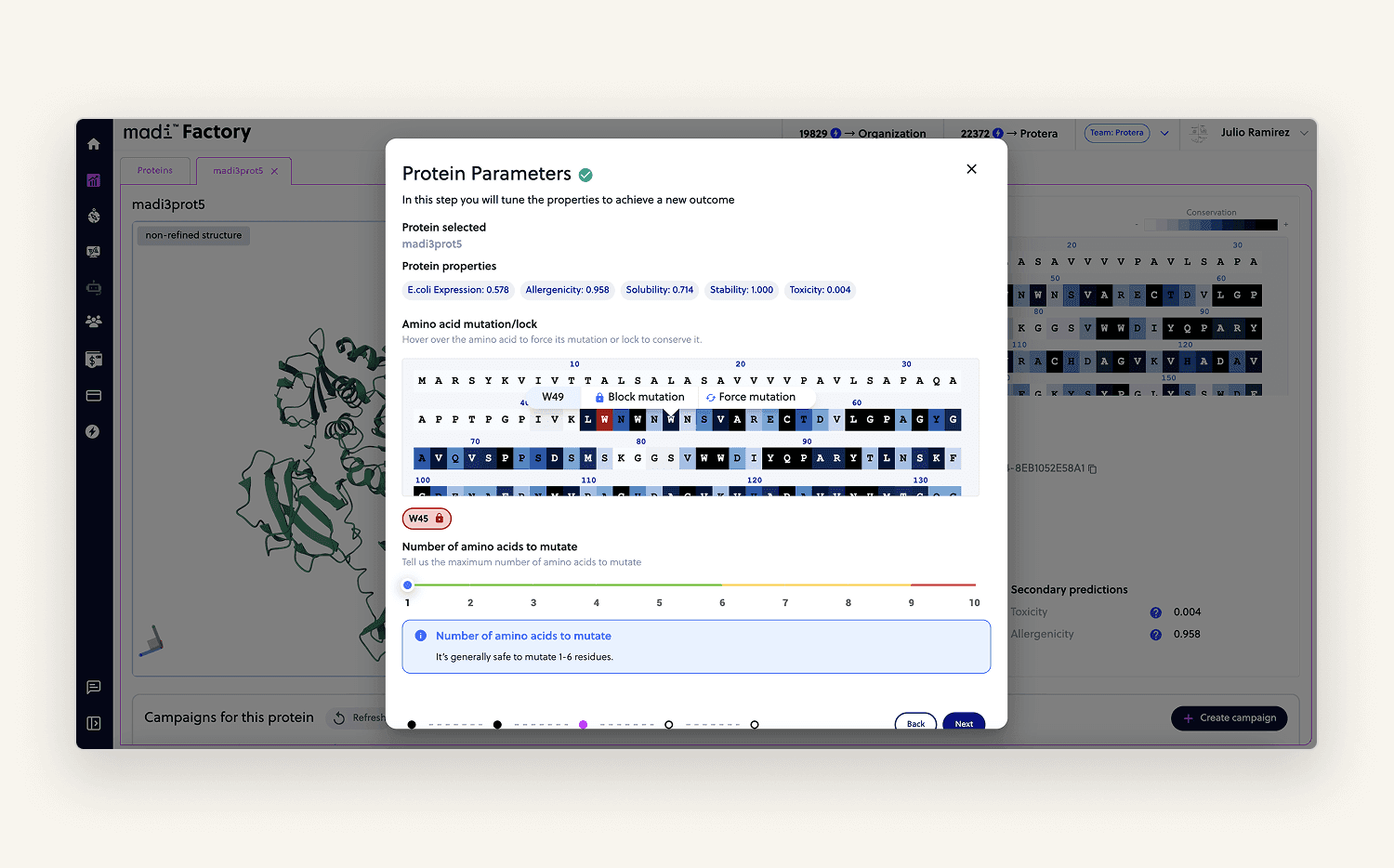
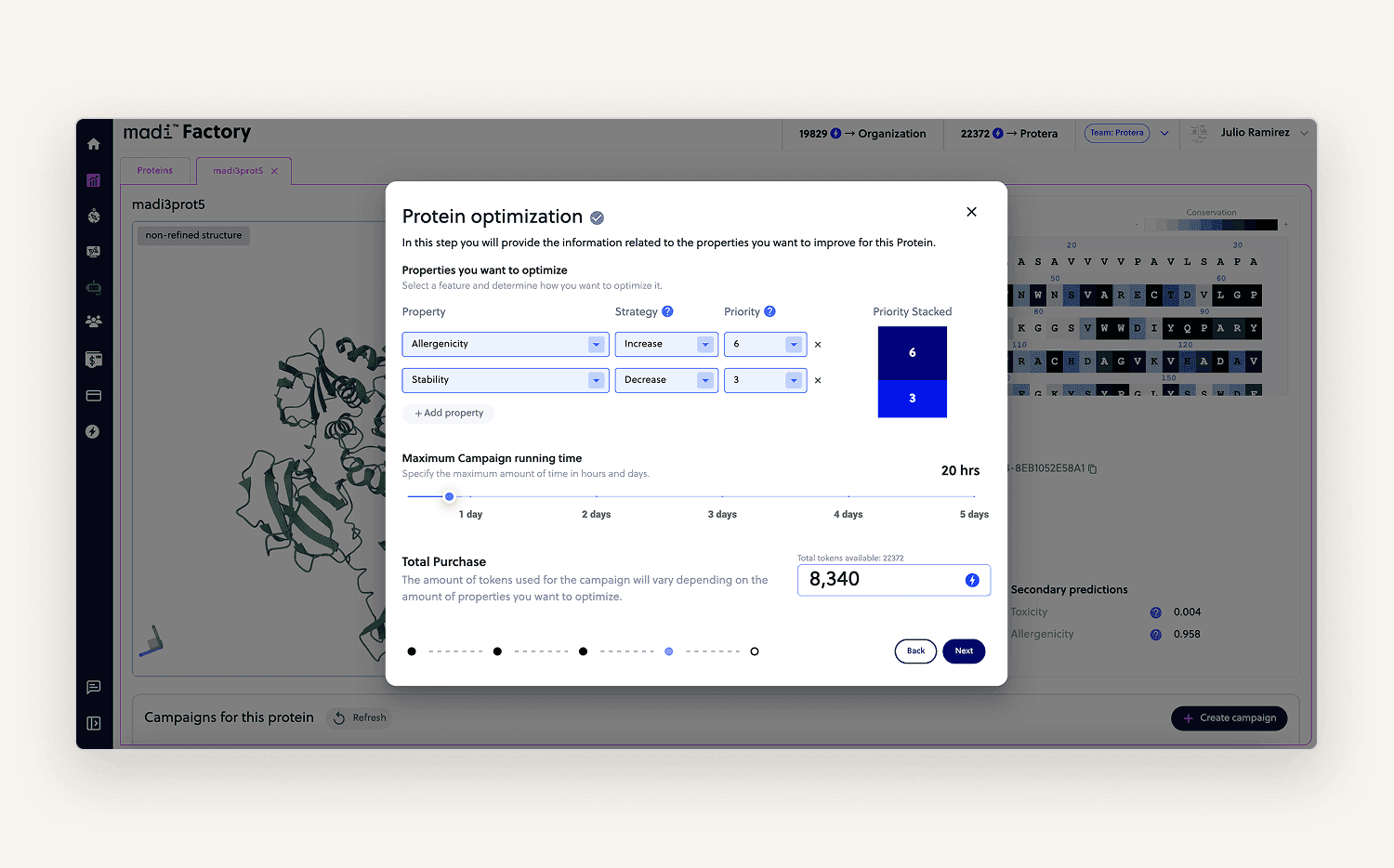
I built a step-by-step flow where users could lock or modify specific amino acids giving the model the right context for optimization. The view also included calculated properties and a 3D rendering (PDB format) of the uploaded protein.
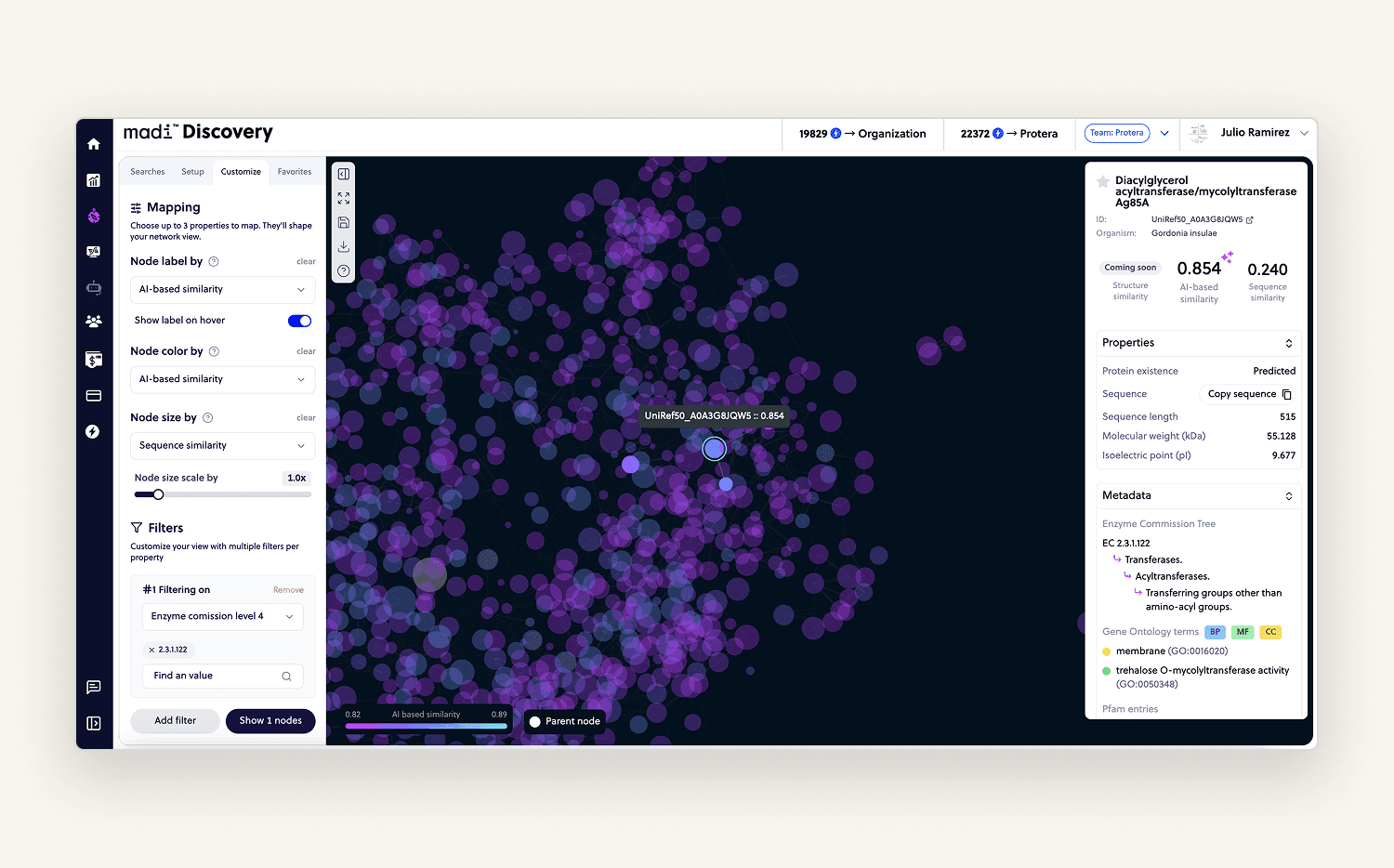
Protein Discovery
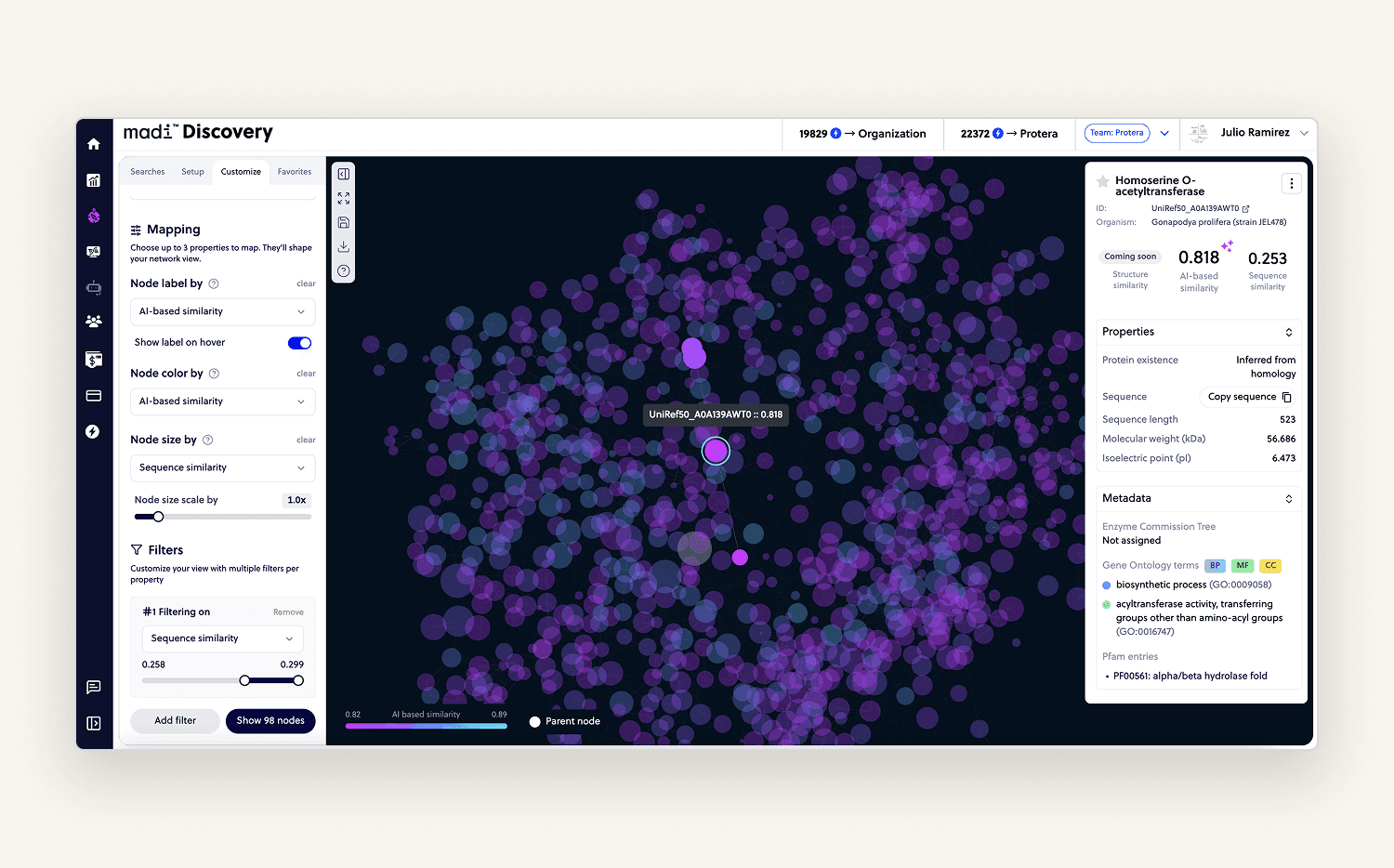
Probably my favorite module to work on. Given one protein, users could explore similar ones from massive public datasets. Results were visualized as an interactive graph that users could zoom, filter, sort, and navigate.
Clicking a node opened a sidebar with relevant data. Users loved it it helped them make smarter decisions, explore alternatives faster, and saved them weeks of manual research.

Gen Optimization
This was a smaller, more experimental feature. It let users upload genes and receive optimized sequences through an internal tool. The feature was relatively simple, so we were able to launch it quickly and gather feedback right away.
Results
The platform was adopted by global companies like BASF, Bimbo, Genscript, Doosan, and others. It helped them reduce development time and costs dramatically turning months of protein design into just weeks, with data-driven decision-making.
What I learned
This project was one of the most enriching experiences I've had. It challenged me to think beyond frontend code to understand business needs, collaborate with scientists, and propose solutions that respected both science and usability.
If I could do it again, I'd push for stronger structure in documentation and design systems early on. But honestly, that's part of building inside a startup especially in a complex domain like biotech.
What really stayed with me was the feeling of building something meaningful from scratch, alongside people who believed in it just as much.
